Liver Cell Atlas
About
On this website you will find the different published liver single cell RNA-sequencing datasets generated in the labs of Charlotte Scott and Martin Guilliams from the VIB-UGent Centre for Inflammation Research, Belgium. Here you will be able to fully interrogate the datasets for expression of your favourite genes or proteins (when CITE-seq was performed). To search for a gene simply enter the official gene name. To search for a protein please click here to see the specific names to use for the different markers included.
Datasets
Murine MAFLD/NASH
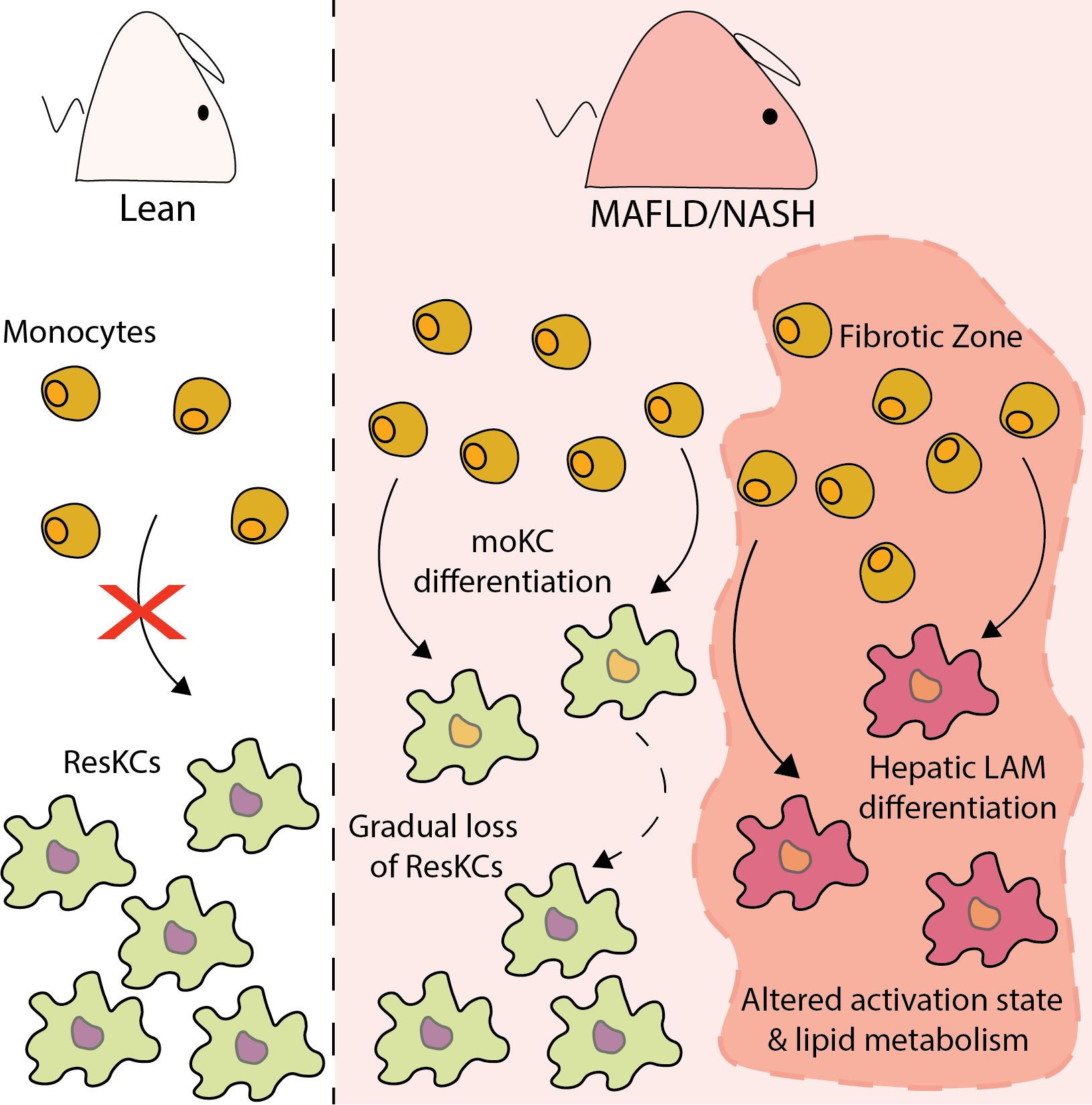
In this publication we have performed CITE-seq analysis on total CD45+ cells isolated from the livers of mice fed a standard diet (SD) or western diet (WD; fat, cholesterol and sugar) for 12, 24 and 36 weeks. In addition, we also zoomed in on the monocyte and macrophage clusters in a separate UMAP to be able to more fully interrogate the data. We also performed single cell RNA sequencing analysis of the CD45- cells isolated from the livers of the mice fed the SD or WD for 24 and 36 weeks. All datasets as presented in the publication are available here for interrogation. Genes and proteins can be searched and overall expression in the UMAPs as well as expression per cluster and in specific clusters over the course of the feeding can be displayed.
Osteopontin Expression Identifies a Subsetof Recruited Macrophages Distinctfrom Kupffer Cells in the Fatty Liver. Anneleen Remmerie, Liesbet Martens, Tinne Thoné, Angela Castoldi, Ruth Seurinck, Benjamin Pavie, Joris Roels, Bavo Vanneste, Sofie De Prijck, Mathias Vanhockerhout, Mushida Binte Abdul Latib, Lindsey Devisscher, Anne Hoorens, Johnny Bonnardel, Niels Vandamme, Anna Kremer, Peter Borghgraef, Hans Van Vlierberghe, Saskia Lippens, Edward Pearce, Yvan Saeys, and Charlotte L. Scott. Immunity, 2020.
https://www.cell.com/immunity/fulltext/S1074-7613(20)30357-5Liver Cell Atlas
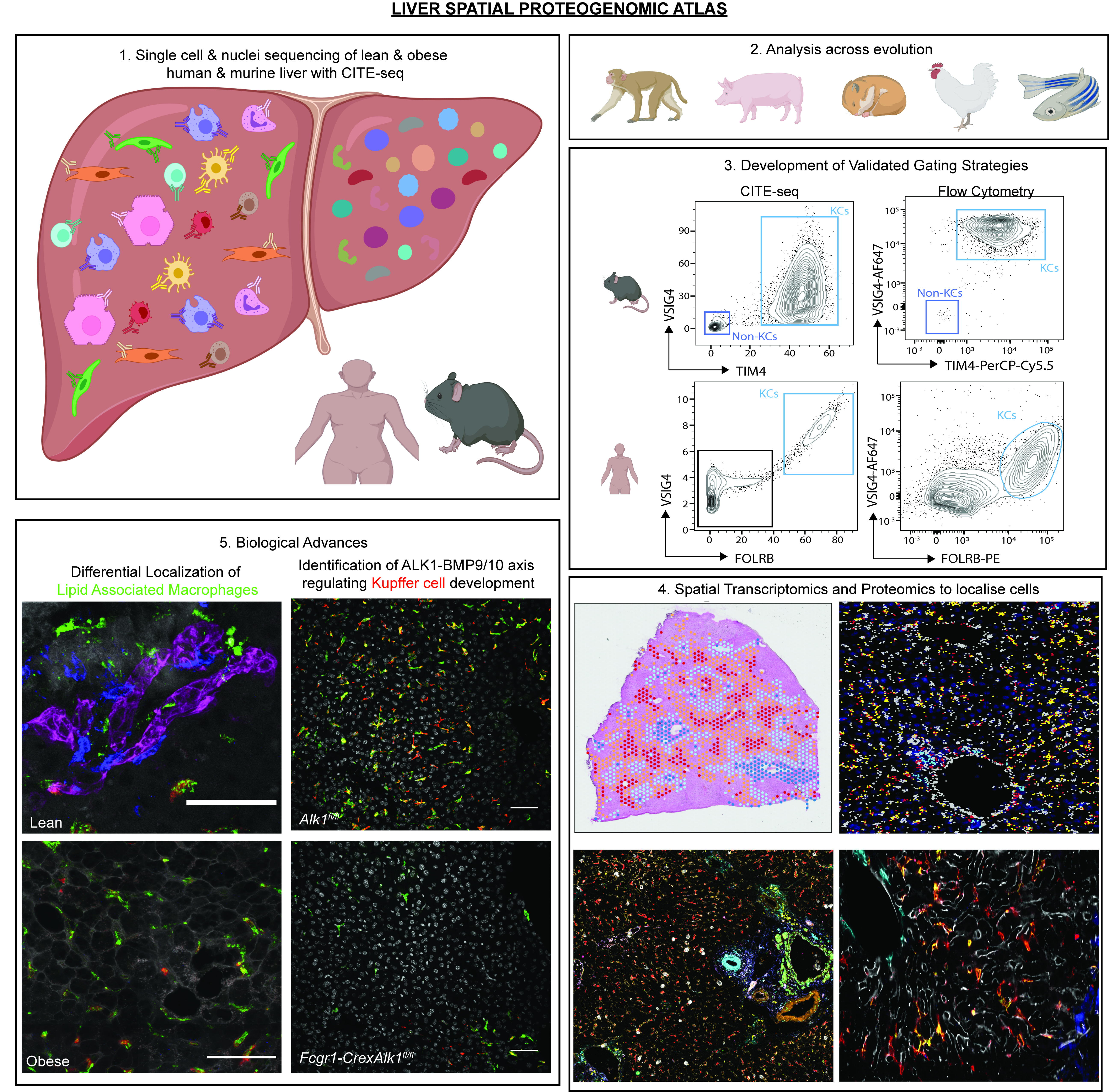
The liver is the largest solid organ in the body, yet it remains incompletely characterized. Here, we present a spatial proteogenomic atlas of the healthy human and murine liver combining single-cell CITE-seq, single-nuclei sequencing, spatial transcriptomics and spatial proteomics. By integrating these multi-omic datasets, we provide validated strategies to reliably discriminate and localize all hepatic cells including a population of bile-duct lipid-associated macrophages (LAMs). We then align this atlas across seven species, revealing the conserved program of bona fide Kupffer cells and bile-duct LAMs. We also uncover the respective spatially-resolved cellular niches of these macrophages and the microenvironmental circuits driving their unique transcriptomic identities. We demonstrate that bile-duct LAMs are induced by local lipid exposure, leading to their induction in steatotic regions of the murine and human liver, while Kupffer cell development crucially depends on their crosstalk with hepatic stellate cells via the evolutionarily-conserved ALK1-BMP9/10 axis.
Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Martin Guilliams, Johnny Bonnardel, Birthe Haest, Bart Vanderborght, Camille Wagner, Anneleen Remmerie, Anna Bujko, Liesbet Martens, Tinne Thoné, Robin Browaeys, Federico F. De Ponti, Bavo Vanneste, Christian Zwicker, Freya R. Svedberg, Tineke Vanhalewyn, Amanda Gonçalves, Saskia Lippens, Bert Devriendt, Eric Cox, Giuliano Ferrero, Valerie Wittamer, Andy Willaert, Suzanne J.F. Kaptein, Johan Neyts, Kai Dallmeier, Peter Geldhof, Stijn Casaert, Bart Deplancke, Peter ten Dijke, Anne Hoorens, Aude Vanlander, Frederik Berrevoet, Yves Van Nieuwenhove, Yvan Saeys, Wouter Saelens, Hans Van Vlierberghe, Lindsey Devisscher and Charlotte L. Scott. Cell, 2022.
https://www.cell.com/cell/fulltext/S0092-8674(21)01481-1Murine APAP/CCL4
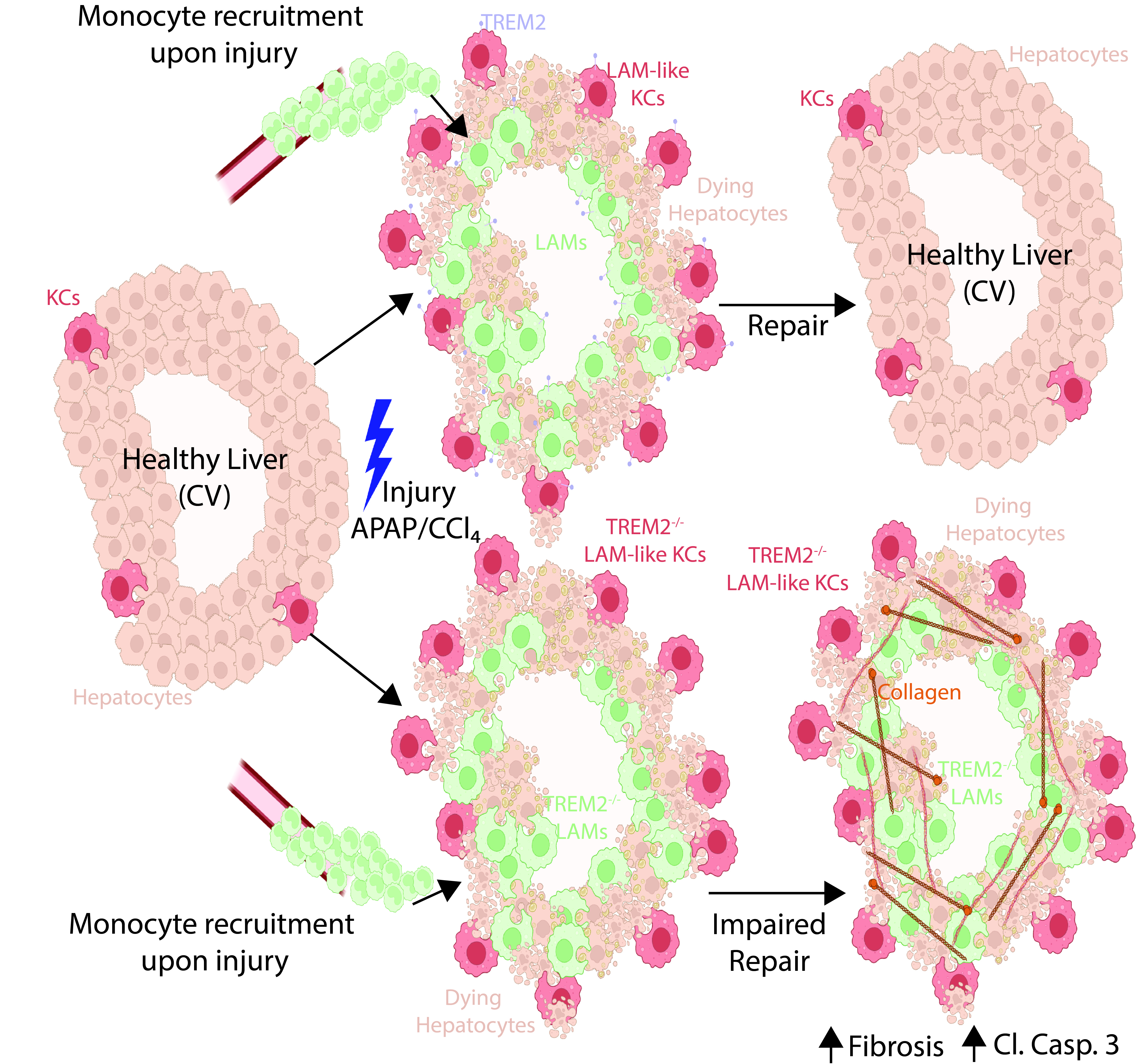
Our understanding of the functional heterogeneity of resident versus recruited macrophages in the diseased liver is limited. A population of recruited lipid associated macrophages (LAMs) has been reported to populate the diseased liver alongside resident Kupffer cells (KCs). However, the precise roles of these distinct macrophage subsets remain elusive. Here, using proteogenomics, we have identified LAMs in multiple models of liver injury. Moreover, we found that this phenotype is not specific to recruited macrophages, as a subset of resident KCs can also adopt a LAM-like phenotype in the mouse and human liver. By combining genetic mouse models targeting the distinct populations, we determined that both recruited LAMs and resident LAM-like KCs play crucial roles in tissue repair. Specifically, triggering receptor expressed on myeloid cells 2 (TREM2) expression on either resident or recruited macrophages is required for the efficient clearance of dying cells, enhancing repair and preventing exacerbated fibrosis.
Spatially-restricted and ontogenically-distinct hepatic macrophages are required for tissue repair. Federico F. De Ponti, Anna Bujko, Zhuangzhuang Liu, Paul J. Collins, Sara Schuermans, Christian Maueroder, Seraja Amstelveen, Tinne Thoné, Liesbet Martens, John G. McKendrick, Pieter A. Louwe, Ana Sànchez Cruz, Wouter Saelens, Kylie P. Matchett, Kathryn J. Waller, Christian Zwicker, Aimée Buglar-Lamb, Bavo Vanneste, Fleur Parmentier, Mushida Binte Abdul Latib, Anneleen Remmerie, Lenard Kertesz, Anneke Kremer, Jérémy Verbeke, David Højland Ipsen, Dominik Reinhard Pfister, Zhaoyuan Liu, Martin Guilliams, Neil C. Henderson, Kodi Ravichandran, Pedro E. Marques, and Charlotte L. Scott. Immunity, 2025.
https://www.cell.com/immunity/fulltext/S1074-7613(25)00002-0Contact
These results were obtained by the teams of Charlotte Scott and Martin Guilliams at the VIB inflammation research center. We hope you will find this resource useful and we would be happy to hear about your experiences and suggestions.
 |
Charlotte Scott, PhD  |
|---|---|
| Professor - Group leader | |
| Laboratory of Myeloid Cell Biology in Tissue Damage and Inflammation | |
| VIB Inflammation Research Center (IRC) | |
| Universiteit Gent | |
| Technologiepark 71, 9052 Zwijnaarde, Belgium | |
| Tel: +32 933 13740 | |
| charlotte.scott@irc.vib-ugent.be |
 |
Martin Guilliams, PhD  |
|---|---|
| Professor - Group leader | |
| Laboratory of Myeloid Cell Biology in Tissue Homeostasis and Regeneration | |
| VIB Inflammation Research Center (IRC) | |
| Universiteit Gent | |
| Technologiepark 71, 9052 Zwijnaarde, Belgium | |
| Tel: +32 933 13740 | |
| martin.guilliams@irc.vib-ugent.be |
| Technical problems | |
|---|---|
| bioit-guilliotts@irc.vib-ugent.be |


